# A tibble: 14 × 2
Data.File.Name Data.File.Description
<chr> <chr>
1 BPX_J Blood Pressure
2 BMX_J Body Measures
3 OHXDEN_J Oral Health - Dentition
4 OHXREF_J Oral Health - Recommendation of…
5 DXXFEM_J Dual-Energy X-ray Absorptiometr…
6 DXX_J Dual-Energy X-ray Absorptiometr…
7 DXXSPN_J Dual-Energy X-ray Absorptiometr…
8 LUX_J Liver Ultrasound Transient Elas…
9 DXXAG_J Dual-Energy X-ray Absorptiometr…
10 BPXO_J Blood Pressure - Oscillometric …
11 AUX_J Audiometry
12 AUXAR_J Audiometry - Acoustic Reflex
13 AUXTYM_J Audiometry - Tympanometry
14 AUXWBR_J Audiometry - Wideband Reflectan…Introduction to Data Linkage
ID 529: Data Management and Analytic Workflows in R
Dean Marengi | Friday, January 13th, 2023
Motivation
- We’ve now discussed:
- Data manipulation using
dplyr - Leveraging
tidyversepackages to work with specific types of data- Dates, date/times, and factors
- Text strings
- Data manipulation using
- However, we’ve only considered data manipulation in the context of individual datasets
- We are often interested in combining data from multiple different files or sources
- Need tools that help us work with and combine multiple datasets
Learning objectives
- Understand the basic principles of data linkage
- The common methods implemented for joining datasets
- How the methods differ, and why these differences should inform which method you implement
- Learn about different R functions available for data linkage
dplyrtwo-table verbs
- Learn how to implement
dplyrtwo-table verbs to link datasets- Mutating joins
- Filtering joins
- Set operations
Background
Why do we need data linkage?
- It’s rare that all the data we need for an analysis are contained within a single file
- Often interested in combining data across two or more files or sources
- For example, NHANES generates a number of datasets for each survey cycle
- Separate files for demographics, clinical exams, laboratory tests, questionnaires, etc.
- Participant data from a given survey cycle must be linked across multiple datasets with a unique identifier
- This is how the
ID529data::nhanes_id529dataset was created!
# A tibble: 10 × 2
Data.File.Name Data.File.Description
<chr> <chr>
1 DR1TOT_J Dietary Interview - Total Nutri…
2 DR2TOT_J Dietary Interview - Total Nutri…
3 DR1IFF_J Dietary Interview - Individual …
4 DR2IFF_J Dietary Interview - Individual …
5 DS1IDS_J Dietary Supplement Use 24-Hour …
6 DSQTOT_J Dietary Supplement Use 30-Day -…
7 DS2IDS_J Dietary Supplement Use 24-Hour …
8 DS1TOT_J Dietary Supplement Use 24-Hour …
9 DS2TOT_J Dietary Supplement Use 24-Hour …
10 DSQIDS_J Dietary Supplement Use 30-Day -…How do we link datasets together in R?
- There are many R functions that allow us to link data
base Rincludes themerge()functiondplyrhas many functions (verbs) for performing two-table operations
- We will focus on
dplyrdplyrfunctions tend to be more consistent, and intuitive to implement- Can be combined into chained data manipulation sequences (
%>%) - Also offers additional types of joins such as semi-joins and anti-joins
dplyr two table verbs
Three classes of dplyr verbs that work with two tables at a time
- Mutating joins
- Combine variables from multiple tables
- Adds new columns to one table with matched rows from another table
- Two types:
- Inner join:
inner_join() - Outer joins:
left_join(),right_join(),full_join()
- Inner join:
- Filtering joins
- Filter rows from one table if they match a row in a second table
- Unmatched rows are discarded if join condition is not met
- Great tools for identifying row mismatches between tables
semi_join(),anti_join
- Filter rows from one table if they match a row in a second table

dplyr two table verbs (cont.)
- Set operations
- Combine observations from two datasets, but treats observations as set elements
intersect(),union(),setdiff()

Example dataset
Overview
NHANESdataset available on the ID529 GitHub- The dataset includes individual-level:
- Demographic and clinical characteristics
- Socioeconomic parameters
- Blood measures of PFAS/PFOA
- Dietary intake parameters
- For our examples, we will split these data into smaller datasets that each contain different variables, identifiers, and number of observations.
- clinical:
id1,age,race_eth,sbp,ht,wt - pfas:
id1,pfos,pfoa,pfna,pfhs,pfde
- clinical:
# Create demog/clincal characteristics table (id1)
clinical <- nhanes %>%
rename(race_eth = race_ethnicity,
sbp = mean_BP,
ht = height,
wt = weight) %>%
select(id1, age, race_eth, sbp:ht)
# Create pfas data table (id1)
pfas <- nhanes %>%
select(id1, PFOS:PFDE) %>%
rename_with(~ str_to_lower(.)) %>%
filter(rowSums(is.na(.)) < 5)Example dataset
# A tibble: 2,339 × 6
id1 age race_eth sbp wt ht
<chr> <int> <fct> <dbl> <dbl> <dbl>
1 73568 26 Non-Hispanic White 105. 47.1 152.
2 73571 76 Non-Hispanic White 126 102. 172.
3 73574 33 <NA> 121. 56.8 158
4 73576 16 Non-Hispanic Black 109. 67.3 170.
5 73577 32 Hispanic 119. 79.7 166.
6 73578 18 Hispanic 123. 109. 175.
7 73584 13 Non-Hispanic White 109. 53.1 145.
8 73587 14 <NA> 112 110. 169.
9 73597 50 Non-Hispanic Black NaN 104. 180.
10 73598 20 Hispanic 112 86.7 165
# … with 2,329 more rows# A tibble: 2,168 × 6
id1 pfos pfoa pfna pfhs pfde
<chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 73568 2.2 3 0.5 3 0.2
2 73571 10.2 4.77 1.3 2 0.3
3 73574 NA NA 0.7 0.2 0.1
4 73576 4.7 2.37 0.6 7.6 0.2
5 73577 3 1.47 0.4 1.2 0.1
6 73584 7 2.37 0.8 0.8 0.2
7 73587 35.5 6.17 3.3 6.3 1.7
8 73598 4.7 1.8 0.5 1.6 0.2
9 73599 4.5 1.87 1.7 0.8 0.2
10 73600 6.3 1.67 0.5 1.6 0.2
# … with 2,158 more rowsdplyr mutating joins
dplyr::left_join()
Main arguments
x= Data frame 1 (left table)y= Data frame 2 (right table)by= Column names(s) of variables to match rows by (key)suffix= Character string appended to column names that appear in bothxandykeep= IfTRUE, preserve the join keys from bothxandy
Description
- Adds columns from
y(right table) tox(left table) - All rows from
xare preserved in the join- i.e., keeps all rows from
xregardless of whether there’s a match iny
- i.e., keeps all rows from
- Of the joins, used most frequently
- Allow you to add new variables from other tables without dropping observations
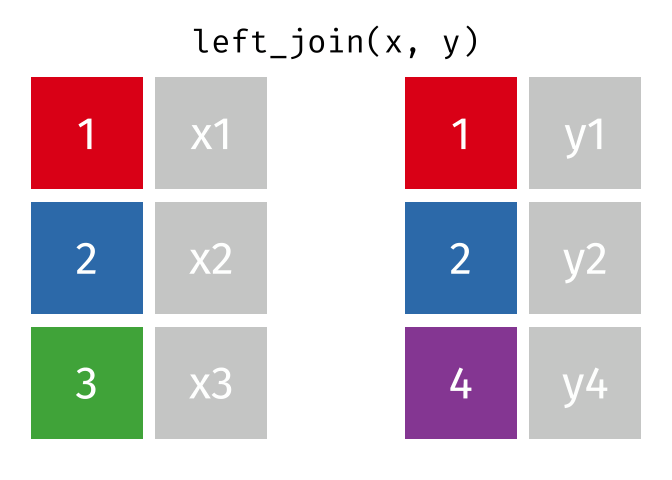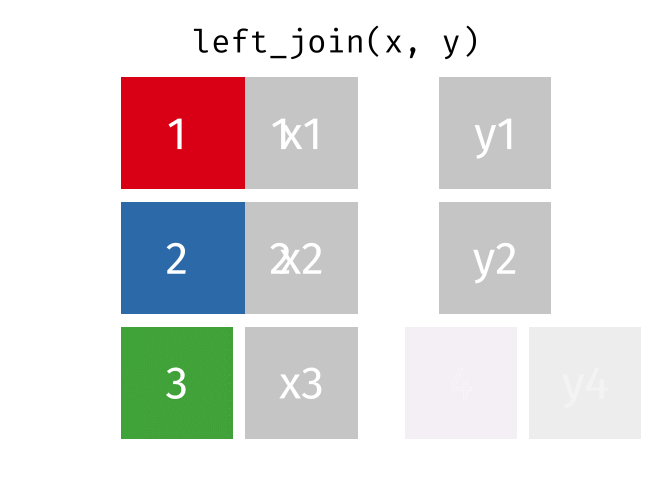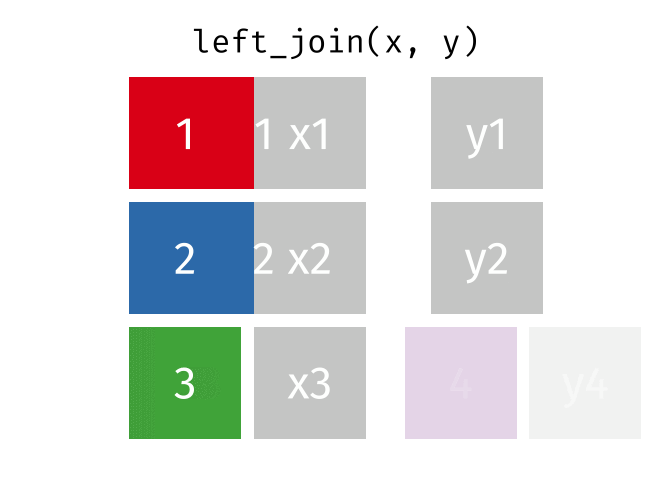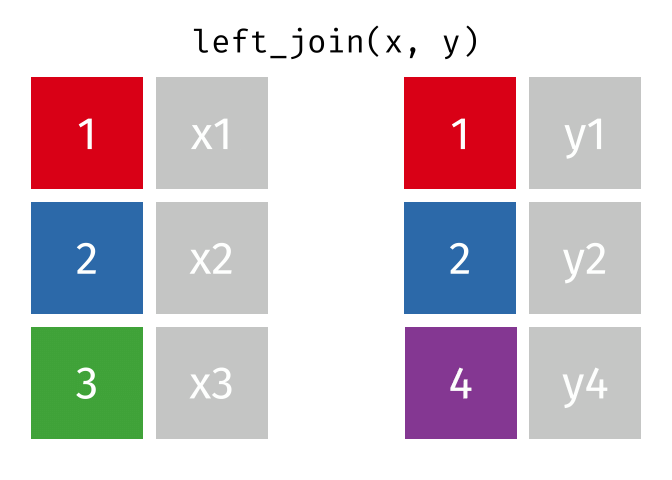
dplyr::left_join() example
# A tibble: 2,339 × 6
id1 age race_eth sbp wt ht
<chr> <int> <fct> <dbl> <dbl> <dbl>
1 73568 26 Non-Hispanic White 105. 47.1 152.
2 73571 76 Non-Hispanic White 126 102. 172.
3 73574 33 <NA> 121. 56.8 158
4 73576 16 Non-Hispanic Black 109. 67.3 170.
5 73577 32 Hispanic 119. 79.7 166.
6 73578 18 Hispanic 123. 109. 175.
7 73584 13 Non-Hispanic White 109. 53.1 145.
8 73587 14 <NA> 112 110. 169.
9 73597 50 Non-Hispanic Black NaN 104. 180.
10 73598 20 Hispanic 112 86.7 165
# … with 2,329 more rows# A tibble: 2,168 × 6
id1 pfos pfoa pfna pfhs pfde
<chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 73568 2.2 3 0.5 3 0.2
2 73571 10.2 4.77 1.3 2 0.3
3 73574 NA NA 0.7 0.2 0.1
4 73576 4.7 2.37 0.6 7.6 0.2
5 73577 3 1.47 0.4 1.2 0.1
6 73584 7 2.37 0.8 0.8 0.2
7 73587 35.5 6.17 3.3 6.3 1.7
8 73598 4.7 1.8 0.5 1.6 0.2
9 73599 4.5 1.87 1.7 0.8 0.2
10 73600 6.3 1.67 0.5 1.6 0.2
# … with 2,158 more rows# A tibble: 2,339 × 11
id1 age race_eth sbp wt ht pfos
<chr> <int> <fct> <dbl> <dbl> <dbl> <dbl>
1 73568 26 Non-Hispan… 105. 47.1 152. 2.2
2 73571 76 Non-Hispan… 126 102. 172. 10.2
3 73574 33 <NA> 121. 56.8 158 NA
4 73576 16 Non-Hispan… 109. 67.3 170. 4.7
5 73577 32 Hispanic 119. 79.7 166. 3
6 73578 18 Hispanic 123. 109. 175. NA
7 73584 13 Non-Hispan… 109. 53.1 145. 7
8 73587 14 <NA> 112 110. 169. 35.5
9 73597 50 Non-Hispan… NaN 104. 180. NA
10 73598 20 Hispanic 112 86.7 165 4.7
# … with 2,329 more rows, and 4 more variables:
# pfoa <dbl>, pfna <dbl>, pfhs <dbl>,
# pfde <dbl>dplyr::right_join()
Main arguments
x= Data frame 1 (left table)y= Data frame 2 (right table)by= Column names(s) of variables to match rows by (key)suffix= Character string appended to column names that appear in bothxandykeep= IfTRUE, preserve the join keys from bothxandy
Description
- Adds columns from
x(left table) toy(right table) - All rows from
yare preserved in the join- i.e., keeps all rows from
yregardless of whether there’s a match inx
- i.e., keeps all rows from
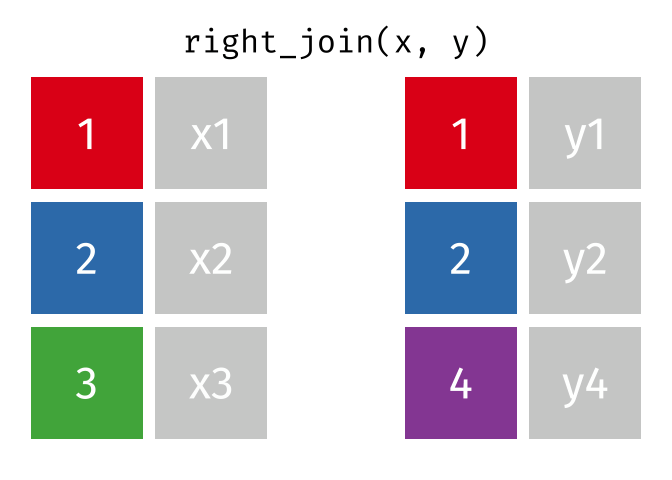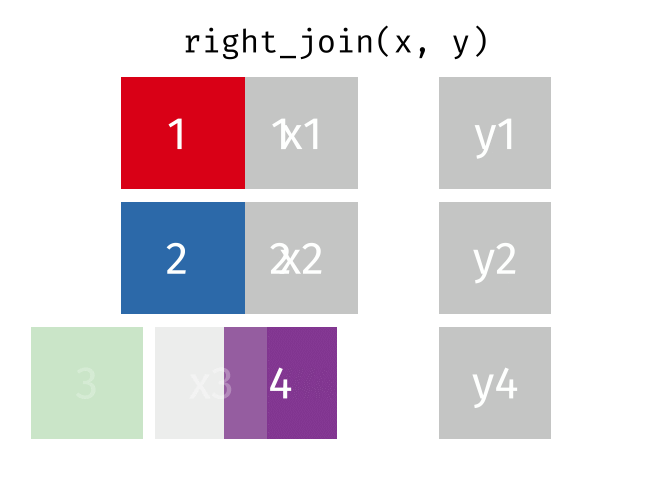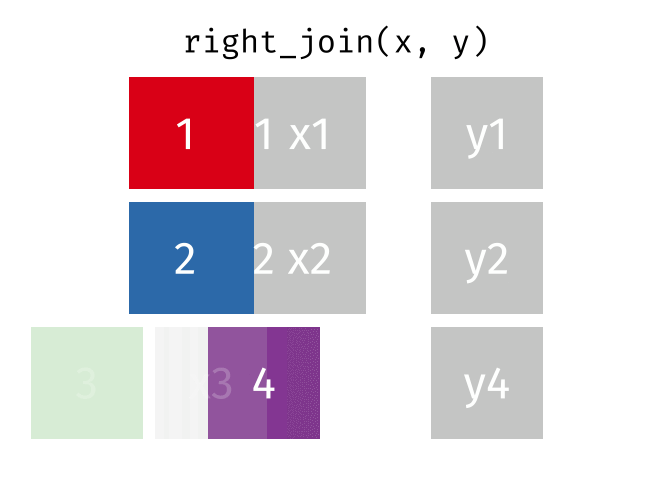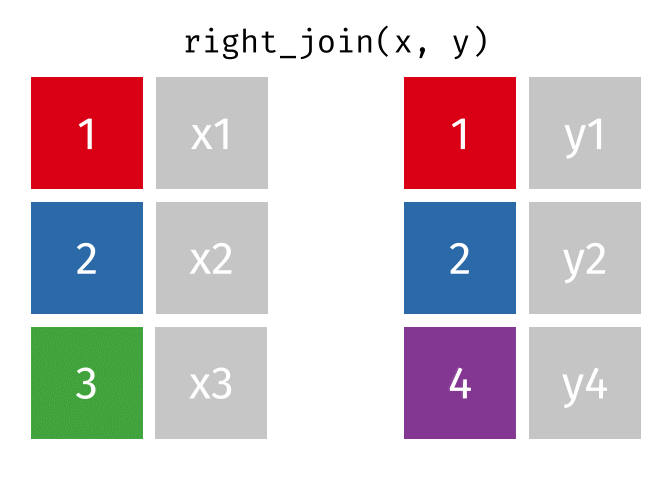
dplyr::right_join() example
# A tibble: 2,339 × 6
id1 age race_eth sbp wt ht
<chr> <int> <fct> <dbl> <dbl> <dbl>
1 73568 26 Non-Hispanic White 105. 47.1 152.
2 73571 76 Non-Hispanic White 126 102. 172.
3 73574 33 <NA> 121. 56.8 158
4 73576 16 Non-Hispanic Black 109. 67.3 170.
5 73577 32 Hispanic 119. 79.7 166.
6 73578 18 Hispanic 123. 109. 175.
7 73584 13 Non-Hispanic White 109. 53.1 145.
8 73587 14 <NA> 112 110. 169.
9 73597 50 Non-Hispanic Black NaN 104. 180.
10 73598 20 Hispanic 112 86.7 165
# … with 2,329 more rows# A tibble: 2,168 × 6
id1 pfos pfoa pfna pfhs pfde
<chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 73568 2.2 3 0.5 3 0.2
2 73571 10.2 4.77 1.3 2 0.3
3 73574 NA NA 0.7 0.2 0.1
4 73576 4.7 2.37 0.6 7.6 0.2
5 73577 3 1.47 0.4 1.2 0.1
6 73584 7 2.37 0.8 0.8 0.2
7 73587 35.5 6.17 3.3 6.3 1.7
8 73598 4.7 1.8 0.5 1.6 0.2
9 73599 4.5 1.87 1.7 0.8 0.2
10 73600 6.3 1.67 0.5 1.6 0.2
# … with 2,158 more rows# A tibble: 2,168 × 11
id1 age race_eth sbp wt ht pfos
<chr> <int> <fct> <dbl> <dbl> <dbl> <dbl>
1 73568 26 Non-Hispan… 105. 47.1 152. 2.2
2 73571 76 Non-Hispan… 126 102. 172. 10.2
3 73574 33 <NA> 121. 56.8 158 NA
4 73576 16 Non-Hispan… 109. 67.3 170. 4.7
5 73577 32 Hispanic 119. 79.7 166. 3
6 73584 13 Non-Hispan… 109. 53.1 145. 7
7 73587 14 <NA> 112 110. 169. 35.5
8 73598 20 Hispanic 112 86.7 165 4.7
9 73599 13 Non-Hispan… 118 44.9 159. 4.5
10 73600 37 Non-Hispan… 149. 126. 185. 6.3
# … with 2,158 more rows, and 4 more variables:
# pfoa <dbl>, pfna <dbl>, pfhs <dbl>,
# pfde <dbl>dplyr::inner_join()
Main arguments
x= Data framexy= Data frameyby= Column name(s) to joinxandybysuffix= Suffix to append to duplicate varskeep= Keep the join key from bothxandy
Description
- Returns rows that match in both
xandy- Unmatched rows are dropped
- Most restrictive of the mutating joins
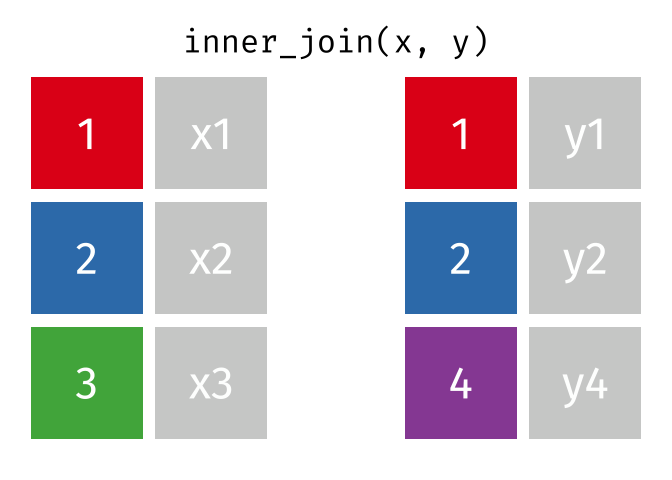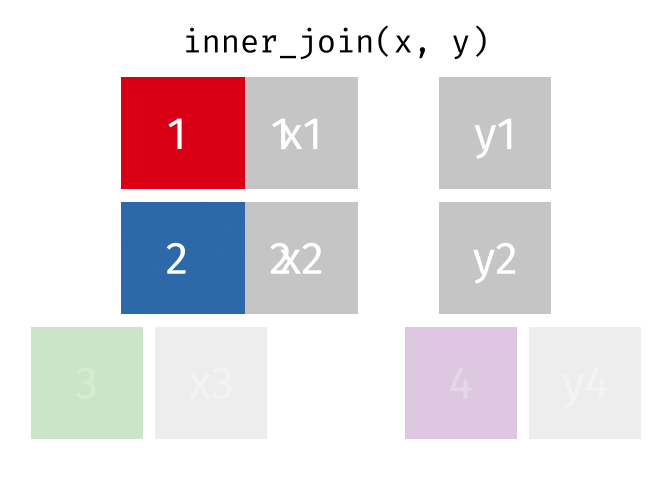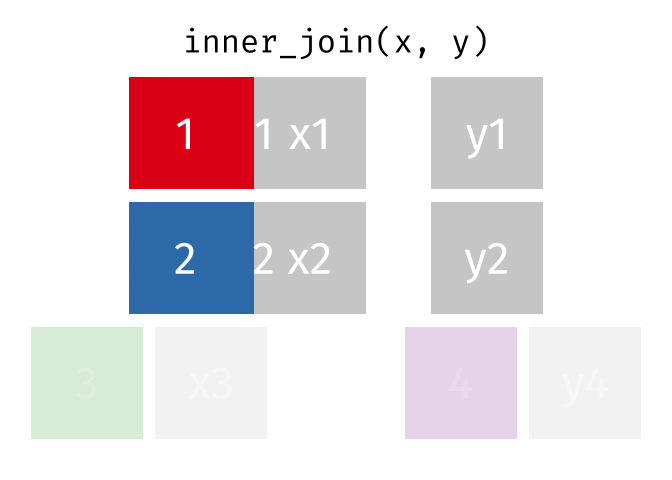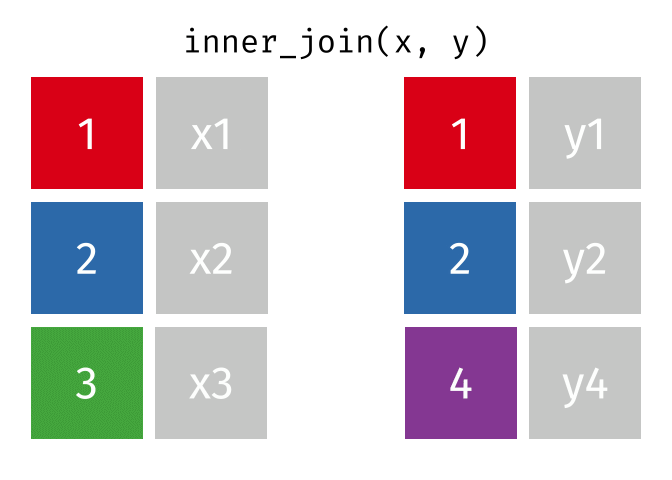
dplyr::inner_join() example
# A tibble: 2,339 × 6
id1 age race_eth sbp wt ht
<chr> <int> <fct> <dbl> <dbl> <dbl>
1 73568 26 Non-Hispanic White 105. 47.1 152.
2 73571 76 Non-Hispanic White 126 102. 172.
3 73574 33 <NA> 121. 56.8 158
4 73576 16 Non-Hispanic Black 109. 67.3 170.
5 73577 32 Hispanic 119. 79.7 166.
6 73578 18 Hispanic 123. 109. 175.
7 73584 13 Non-Hispanic White 109. 53.1 145.
8 73587 14 <NA> 112 110. 169.
9 73597 50 Non-Hispanic Black NaN 104. 180.
10 73598 20 Hispanic 112 86.7 165
# … with 2,329 more rows# A tibble: 2,168 × 6
id1 pfos pfoa pfna pfhs pfde
<chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 73568 2.2 3 0.5 3 0.2
2 73571 10.2 4.77 1.3 2 0.3
3 73574 NA NA 0.7 0.2 0.1
4 73576 4.7 2.37 0.6 7.6 0.2
5 73577 3 1.47 0.4 1.2 0.1
6 73584 7 2.37 0.8 0.8 0.2
7 73587 35.5 6.17 3.3 6.3 1.7
8 73598 4.7 1.8 0.5 1.6 0.2
9 73599 4.5 1.87 1.7 0.8 0.2
10 73600 6.3 1.67 0.5 1.6 0.2
# … with 2,158 more rows# A tibble: 2,168 × 11
id1 age race_eth sbp wt ht pfos
<chr> <int> <fct> <dbl> <dbl> <dbl> <dbl>
1 73568 26 Non-Hispan… 105. 47.1 152. 2.2
2 73571 76 Non-Hispan… 126 102. 172. 10.2
3 73574 33 <NA> 121. 56.8 158 NA
4 73576 16 Non-Hispan… 109. 67.3 170. 4.7
5 73577 32 Hispanic 119. 79.7 166. 3
6 73584 13 Non-Hispan… 109. 53.1 145. 7
7 73587 14 <NA> 112 110. 169. 35.5
8 73598 20 Hispanic 112 86.7 165 4.7
9 73599 13 Non-Hispan… 118 44.9 159. 4.5
10 73600 37 Non-Hispan… 149. 126. 185. 6.3
# … with 2,158 more rows, and 4 more variables:
# pfoa <dbl>, pfna <dbl>, pfhs <dbl>,
# pfde <dbl>dplyr::full_join()
Main arguments
x= Data framexy= Data frameyby= Column name(s) to joinxandybysuffix= Suffix to append to duplicate varskeep= Keep the join key from bothxandy
Description
- Returns all rows in x or y
- Least restrictive
- No observations are removed
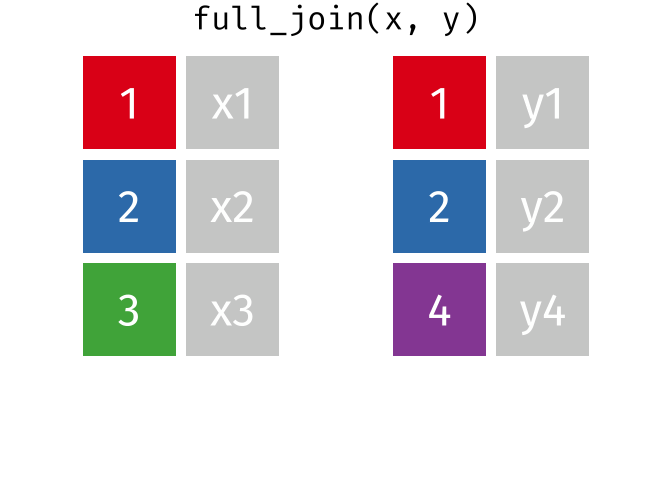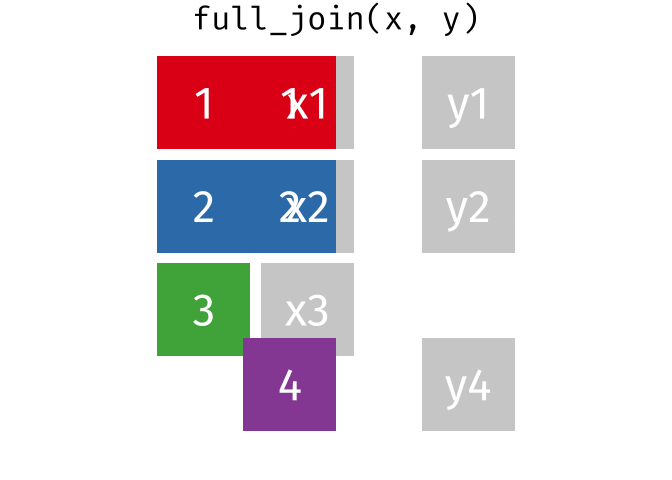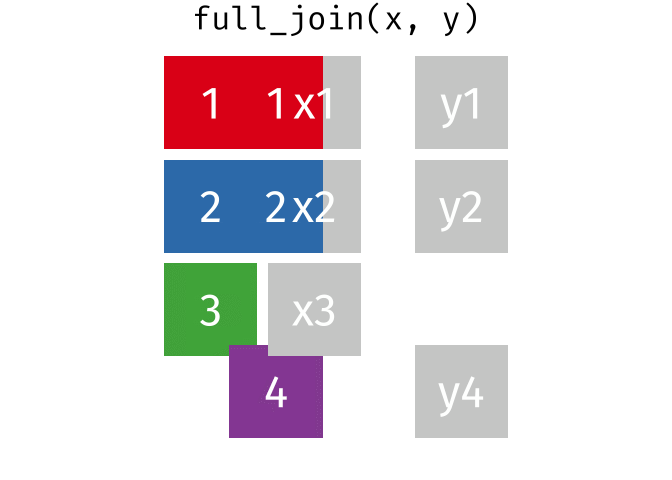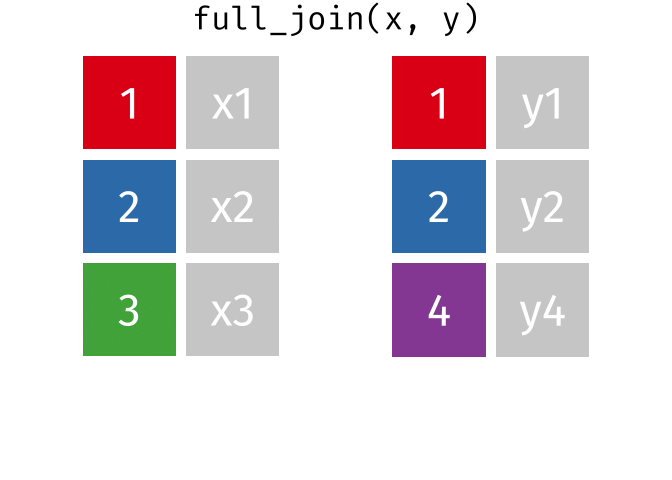
dplyr::full_join() example
# A tibble: 2,339 × 6
id1 age race_eth sbp wt ht
<chr> <int> <fct> <dbl> <dbl> <dbl>
1 73568 26 Non-Hispanic White 105. 47.1 152.
2 73571 76 Non-Hispanic White 126 102. 172.
3 73574 33 <NA> 121. 56.8 158
4 73576 16 Non-Hispanic Black 109. 67.3 170.
5 73577 32 Hispanic 119. 79.7 166.
6 73578 18 Hispanic 123. 109. 175.
7 73584 13 Non-Hispanic White 109. 53.1 145.
8 73587 14 <NA> 112 110. 169.
9 73597 50 Non-Hispanic Black NaN 104. 180.
10 73598 20 Hispanic 112 86.7 165
# … with 2,329 more rows# A tibble: 2,168 × 6
id1 pfos pfoa pfna pfhs pfde
<chr> <dbl> <dbl> <dbl> <dbl> <dbl>
1 73568 2.2 3 0.5 3 0.2
2 73571 10.2 4.77 1.3 2 0.3
3 73574 NA NA 0.7 0.2 0.1
4 73576 4.7 2.37 0.6 7.6 0.2
5 73577 3 1.47 0.4 1.2 0.1
6 73584 7 2.37 0.8 0.8 0.2
7 73587 35.5 6.17 3.3 6.3 1.7
8 73598 4.7 1.8 0.5 1.6 0.2
9 73599 4.5 1.87 1.7 0.8 0.2
10 73600 6.3 1.67 0.5 1.6 0.2
# … with 2,158 more rows# A tibble: 2,339 × 11
id1 age race_eth sbp wt ht pfos
<chr> <int> <fct> <dbl> <dbl> <dbl> <dbl>
1 73568 26 Non-Hispan… 105. 47.1 152. 2.2
2 73571 76 Non-Hispan… 126 102. 172. 10.2
3 73574 33 <NA> 121. 56.8 158 NA
4 73576 16 Non-Hispan… 109. 67.3 170. 4.7
5 73577 32 Hispanic 119. 79.7 166. 3
6 73578 18 Hispanic 123. 109. 175. NA
7 73584 13 Non-Hispan… 109. 53.1 145. 7
8 73587 14 <NA> 112 110. 169. 35.5
9 73597 50 Non-Hispan… NaN 104. 180. NA
10 73598 20 Hispanic 112 86.7 165 4.7
# … with 2,329 more rows, and 4 more variables:
# pfoa <dbl>, pfna <dbl>, pfhs <dbl>,
# pfde <dbl>dplyr filtering joins
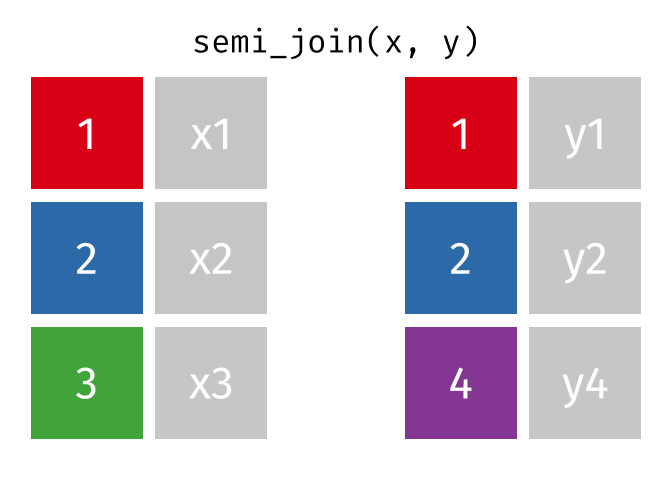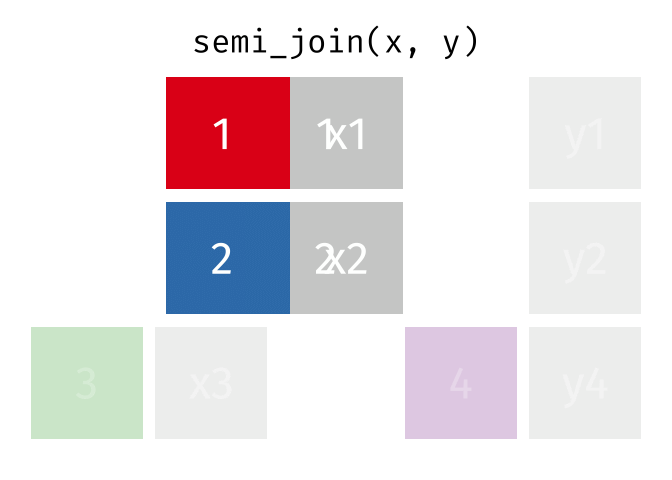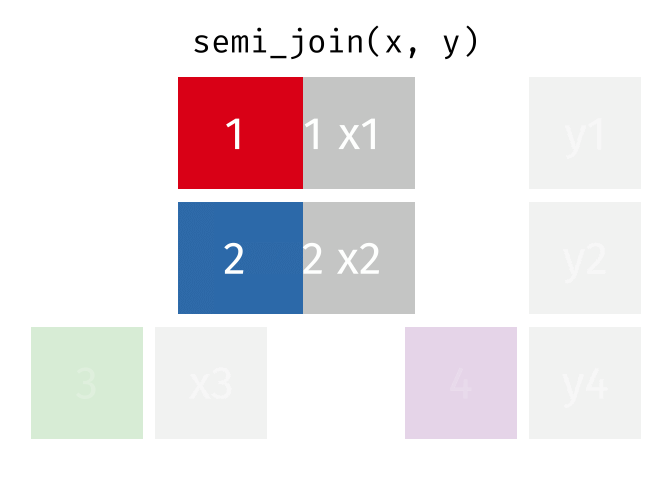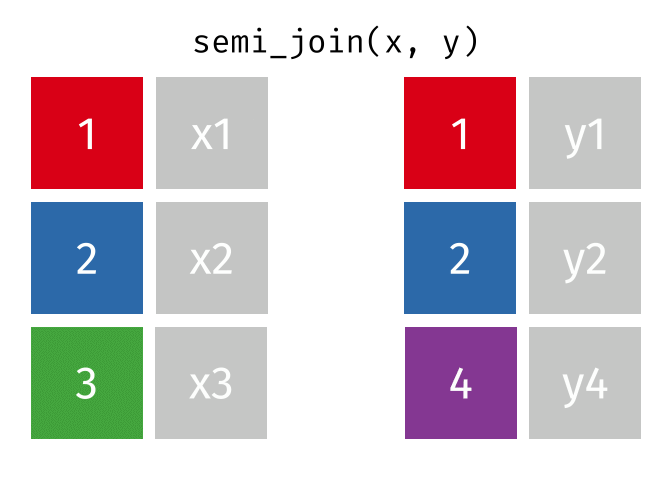
dplyr::semi_join()
Main arguments
x= Data frame 1 (left)y= Data frame 2 (right)by= Column names(s) of variables to match rows by (key)
Description
- Keeps all observations in x that have a match in y
- Useful for filtering
xby the presence of a match iny
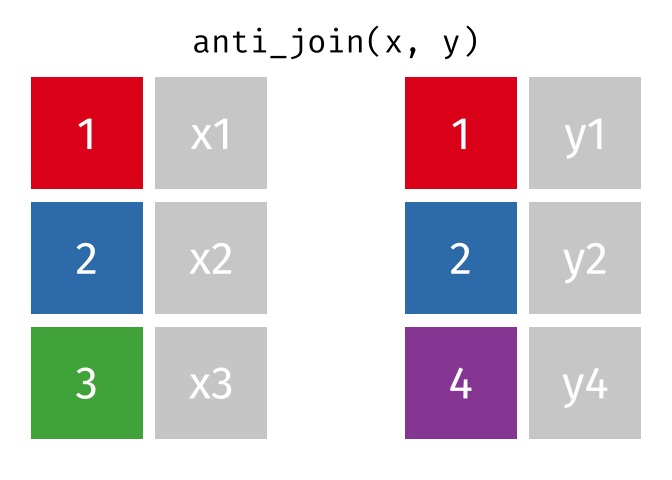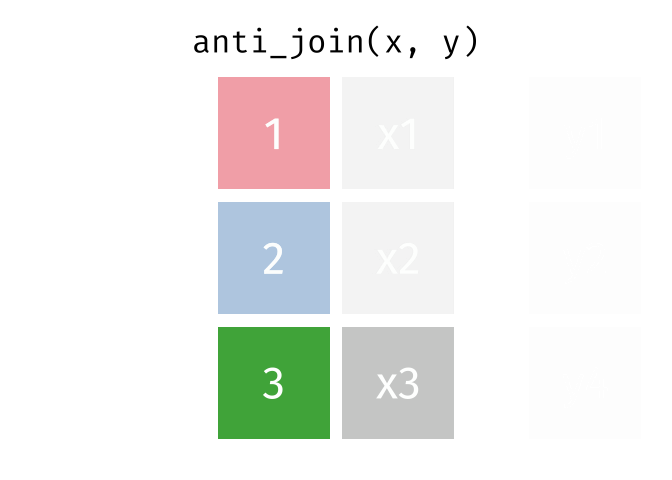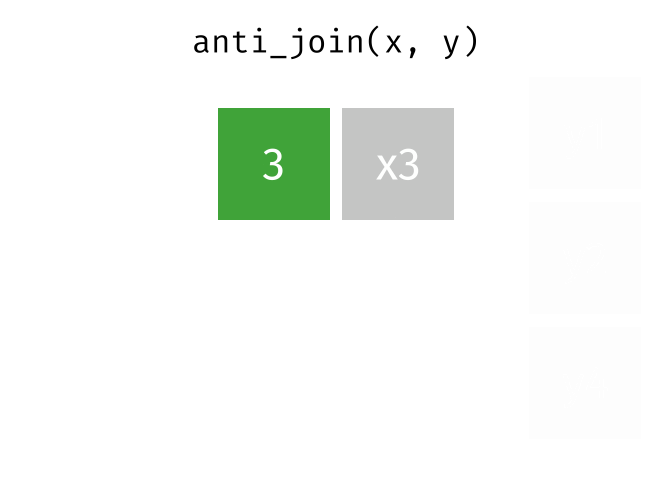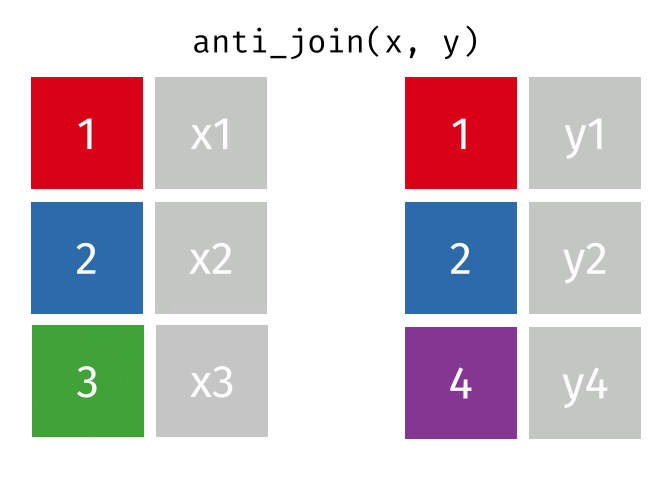
dplyr::anti_join()
Main arguments
x= Data frame 1 (left)y= Data frame 2 (right)by= Column names(s) of variables to match rows by (key)
Description
- Returns rows from
xwhere there is no match iny - Useful when you want to filter the first table by the absence of a match in the second table
- That is, identify observations in
xthat don’t appear iny
- That is, identify observations in
dplyr set operations
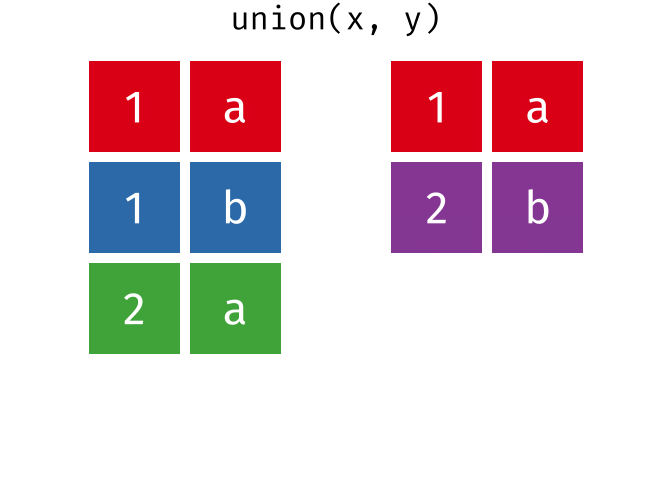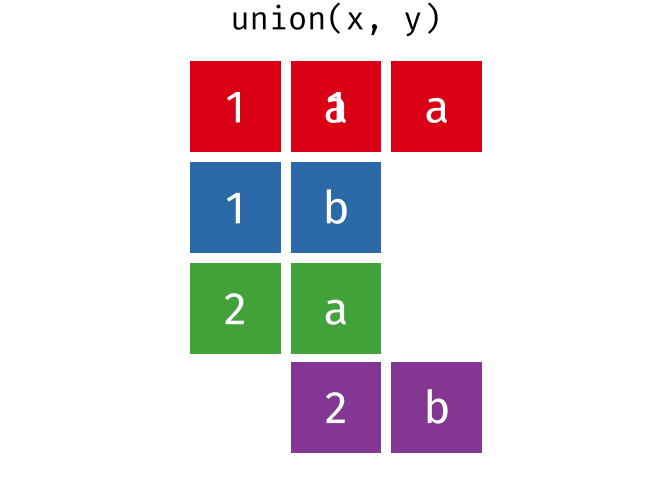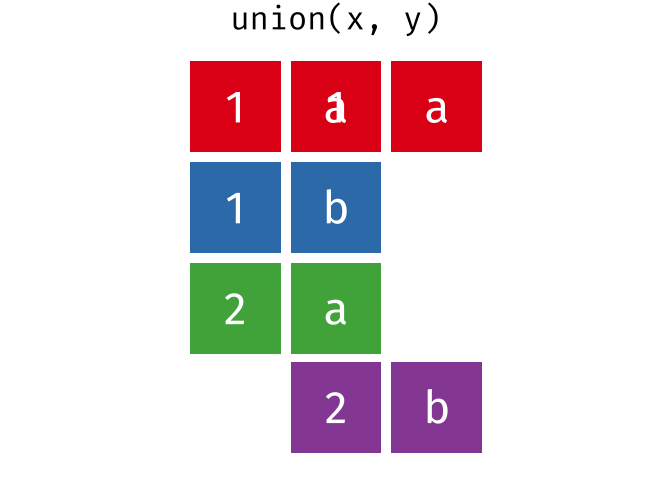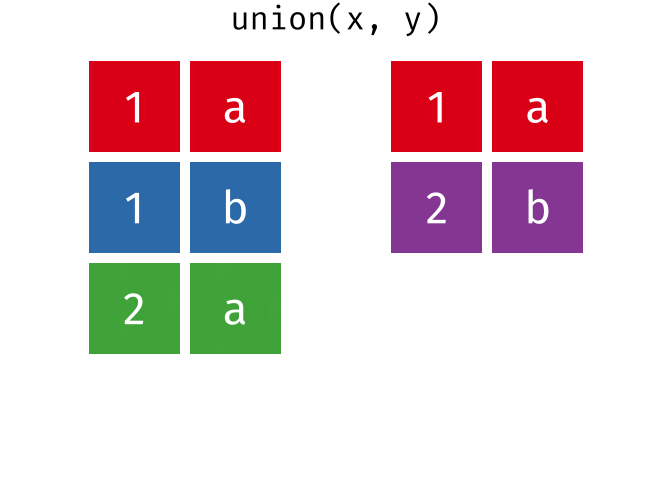
dplyr::union()
Description
union()- Return unique observations in
xandy
- Return unique observations in
union_all()- Return unique and duplicate observations in
xandy
- Return unique and duplicate observations in
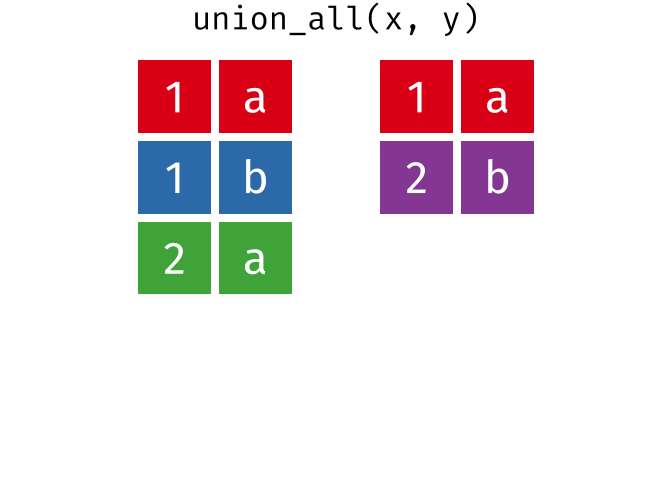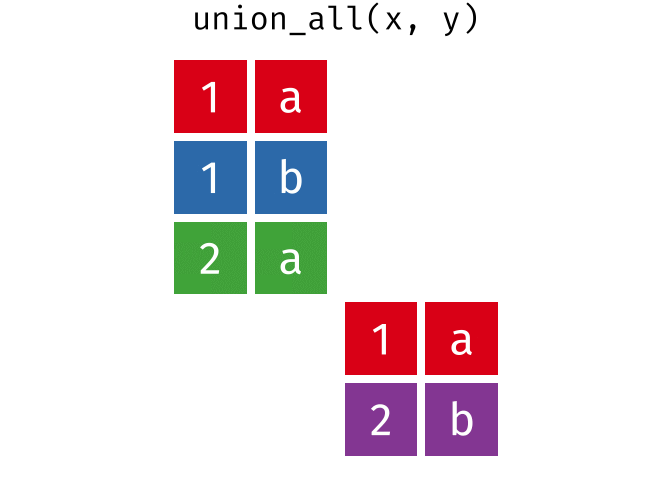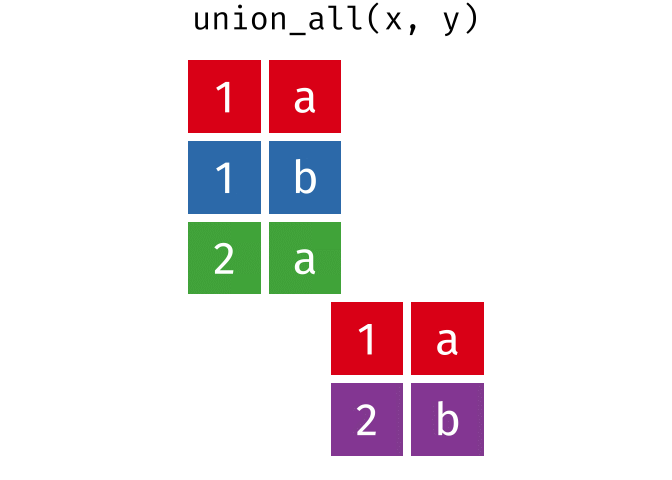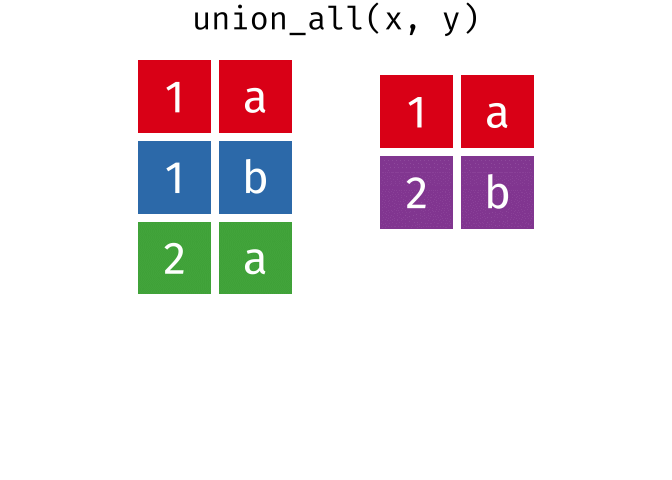
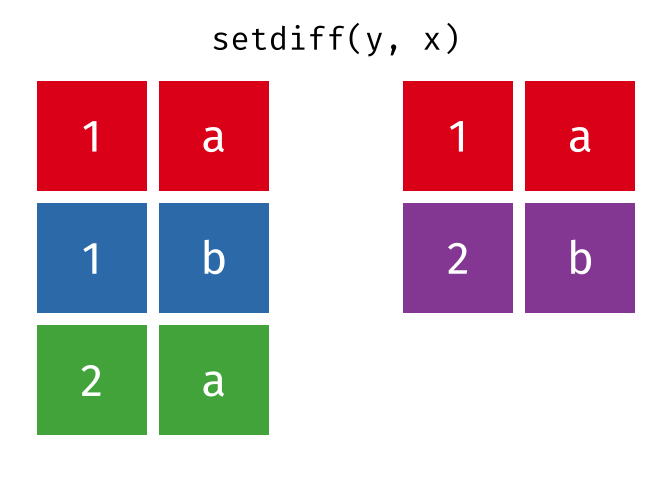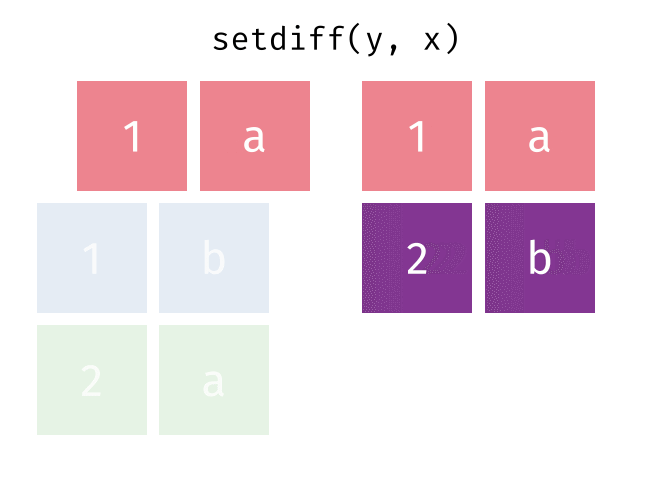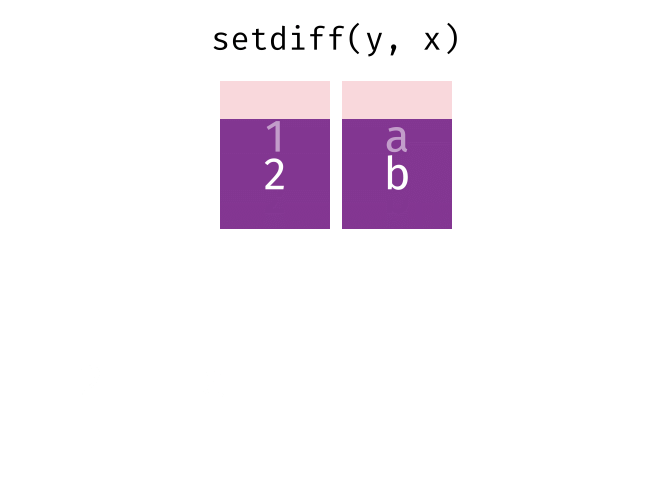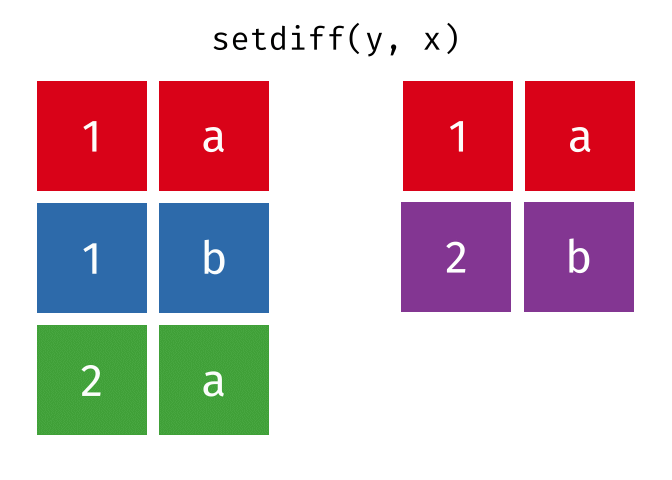
dplyr::setdiff()
Description
- Returns the rows that are in the first table but not in the second
- That is, return observations in x, but not in y
Key takeaways
- Joining together two or more datasets is a routine component of data analysis workflows
- Different types of joins are useful in different situations
- It’s important to be aware of how and when to use each (left, right, inner, etc.)
- All have distinct implications for how the underlying data are manipulated
- Like all data manipulation tasks, joining data can get complicated!
- Common challenges include:
- Inconsistencies in unique identifiers
- Duplicate records and missing values
- Non-unique identifiers (require matching data frames on two or more variables)
- Joining datasets can be complex and requires a good understanding of the data structure
dplyrincludes a number of two-table functions (verbs)- Like other
dplyrverbs, two-table verbs are:- Intuitive to use
- Can be incorporated into a sequence of data cleaning operations using the pipe operator
- Like other
Resources
dplyrTwo-table verbs vignette: https://dplyr.tidyverse.org/articles/two-table.html
- R for Data Science - Relational Data: https://r4ds.had.co.nz/relational-data.html
Activity
- Download R Script file here: https://github.com/ID529/lectures/tree/main/day5/lecture3-data-linkage-methods
nhanes-activity.R
- ID 529 dataset details: https://github.com/ID529/ID529data
