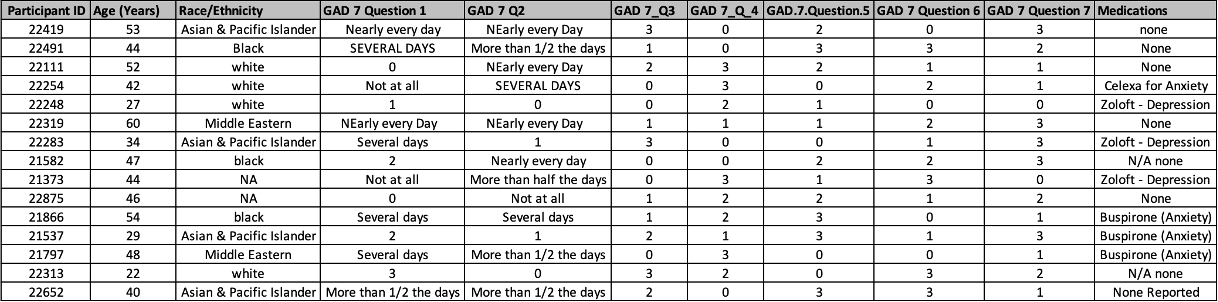
Rows: 15
Columns: 11
$ `Participant ID` <dbl> 22419, 22491, 22111, 22254, 22248, 22319, 22283, 21582, 21373, 22875, 21866, 21537, 21797, 22313, 226…
$ `Age (Years)` <dbl> 53, 44, 52, 42, 27, 60, 34, 47, 44, 46, 54, 29, 48, 22, 40
$ `Race/Ethnicity` <chr> "Asian & Pacific Islander", "Black", "white", "white", "white", "Middle Eastern", "Asian & Pacific Is…
$ `GAD 7 Question 1` <chr> "Nearly every day", "SEVERAL DAYS", "0", "Not at all", "1", "NEarly every Day", "Several days", "2", …
$ `GAD 7 Q2` <chr> "NEarly every Day", "More than 1/2 the days", "NEarly every Day", "SEVERAL DAYS", "0", "NEarly every …
$ `GAD 7_Q3` <dbl> 3, 1, 2, 0, 0, 1, 3, 0, 0, 1, 1, 2, 0, 3, 2
$ `GAD 7_Q_4` <dbl> 0, 0, 3, 3, 2, 1, 0, 0, 3, 2, 2, 1, 3, 2, 0
$ GAD.7.Question.5 <dbl> 2, 3, 2, 0, 1, 1, 0, 2, 1, 2, 3, 3, 0, 0, 3
$ `GAD 7 Question 6` <dbl> 0, 3, 1, 2, 0, 2, 1, 2, 3, 1, 0, 1, 0, 3, 3
$ `GAD 7 Question 7` <dbl> 3, 2, 1, 1, 0, 3, 3, 3, 0, 2, 1, 3, 1, 2, 1
$ Medications <chr> "none", "None", "None", "Celexa for Anxiety", "Zoloft - Depression", "None", "Zoloft - Depression", "…