df <- NHANES |>
# Remember that we have to restrict to people 25 and above
filter(Age>=25) |>
# recoding of the variables we're going to use
mutate(agecat = case_when(
Age < 35 ~ "25-34",
35 <= Age & Age < 45 ~ "35-44",
Age >= 45 & Age < 55 ~ "45-54",
Age >= 55 & Age < 65 ~ "55-64",
Age >= 65 & Age < 75 ~ "65-74",
Age >= 75 ~ "75+"),
# We want College Grad to be the reference category for education, so we'll
# re-order the factor so that it is reversed from the way it came in the NHANES dataset
Education = factor(Education,
levels=rev(levels(NHANES$Education))),
# Here we collapse Hispanic and Mexican into the Hispanic category
racecat = factor(case_when(
Race1 %in% c("Hispanic", "Mexican") ~ "Hispanic",
Race1 %in% c("Asian", "Other") ~ "Other Non-Hispanic",
Race1 == "Black" ~ "Black Non-Hispanic",
Race1 == "White" ~ "White Non-Hispanic"),
levels = c("White Non-Hispanic", "Black Non-Hispanic", "Hispanic", "Other Non-Hispanic"))
) |>
# select just variables we are going to use in the analysis
select(ID, SurveyYr, Gender, Age, agecat, Education, racecat, BPSysAve, SmokeNow)Regression modeling workflows: making it pretty
ID 529: Data Management and Analytic Workflows in R
Jarvis Chen
Friday, January 12, 2024
Follow along
`https://bit.ly/id529_regression_models
Clone the repository and open it as a
RProject in your RStudio.Open the
id529_day4_regression_models.Rscript. Run the data cleaning steps and fit the models on thedf_completecasedata frame
Some goals for today
Learn how to create some pretty tables
- Learn about the
gtsummarypackage - Learn about the
sjPlotpackage - Learn about the
stargazerpackage - Learn how to create plots of regression results
- Learn about the
Data preparation
- As we covered on Day 4, we begin by doing some data cleaning steps.
Restricting to non-missing observations
- Though most regression functions will automatically drop observations with
NA, sometimes we may want to explicitly filter out missing observations on any of the covariates we are going to include while model building to make sure that we can compare models based on the same number of observations.
Fitting multiple models
lm_model1 <- lm(BPSysAve ~ factor(Education),
data=df_completecase)
lm_model2 <- lm(BPSysAve ~ factor(Education) + factor(agecat) + Gender,
data=df_completecase)
lm_model3 <- lm(BPSysAve ~ factor(Education) + factor(agecat) + Gender + factor(racecat),
data=df_completecase)
lm_model4b <- lm(BPSysAve ~ factor(Education) + factor(agecat) + interaction(Gender,factor(racecat)),
data=df_completecase)- Note that in
lm_model4bI’ve used theinteraction()function to create a categorical variable representing the cross-classified categories of gender and race/ethnicity
A workflow for extracting model results into tables
We have been emphasizing the idea of having a workflow that includes not only fitting a statistical model, but
- All of the data cleaning steps that help you get your data set up before you fit one or more statistical models
- All of the extraction and formatting steps that enable you to communicate the results after you fit your statistical model
Given that you will often have to refit models and tweak your analysis many times before presenting your final results, it is helpful to have a workflow that facilitates making edits or revisions and having these propagate to your final tables and figures
gtsummary package
The gtsummary package provides a user-friendly way to create publication-ready analytical and summary tables. It seamlessly integrates with the tidyverse and the gt package, allowing for easy manipulation and styling of tables. Some key functions in the gtsummary package include
tbl_summary(): Creates summary tables for descriptive statistics. It can automatically recognize and handle different data types (e.g., continuous, categorical) and present appropriate summary statistics.tbl_regression(): Used for formatting regression model results. It takes a model object (like those fromlm,glm, etc.) and returns a table of model estimates and statistics, making it easier to report regression analysis findings.tbl_merge(): Allows for merging multiplegtsummarytables into a single table, which is useful for side-by-side comparisons.tbl_stack(): Stacks multiplegtsummarytables vertically, which is helpful when you need to present similar tables for different groups or categories in a consolidated format.add_p(): Adds p-values to the tables, which is particularly useful when summarizing statistical tests.
gtsummary: :tbl_regression( )
gtsummary: :tbl_regression( )
- I don’t like that the table has the label “factor(Education)”, so I can modify this using the
label=argument
tbl_lm_model1 <-
tbl_regression(lm_model1, label = list('factor(Education)' ~ 'Education')) |>
bold_labels()
tbl_lm_model1| Characteristic | Beta | 95% CI1 | p-value |
|---|---|---|---|
| Education | |||
| College Grad | — | — | |
| Some College | 3.8 | 2.7, 4.8 | <0.001 |
| High School | 5.1 | 3.9, 6.4 | <0.001 |
| 9 - 11th Grade | 4.7 | 3.3, 6.1 | <0.001 |
| 8th Grade | 7.9 | 6.1, 9.8 | <0.001 |
| 1 CI = Confidence Interval | |||
gtsummary: :tbl_regression( )
- Note that we can add model fit statistics using
add_glance_table()
tbl_lm_model1_glance <-
tbl_regression(lm_model1, label = list('factor(Education)' ~ 'Education')) |>
bold_labels() |>
add_glance_table()
tbl_lm_model1_glance| Characteristic | Beta | 95% CI1 | p-value |
|---|---|---|---|
| Education | |||
| College Grad | — | — | |
| Some College | 3.8 | 2.7, 4.8 | <0.001 |
| High School | 5.1 | 3.9, 6.4 | <0.001 |
| 9 - 11th Grade | 4.7 | 3.3, 6.1 | <0.001 |
| 8th Grade | 7.9 | 6.1, 9.8 | <0.001 |
| R² | 0.019 | ||
| Adjusted R² | 0.018 | ||
| Sigma | 17.3 | ||
| Statistic | 30.4 | ||
| p-value | <0.001 | ||
| df | 4 | ||
| Log-likelihood | -26,991 | ||
| AIC | 53,994 | ||
| BIC | 54,035 | ||
| Deviance | 1,886,669 | ||
| Residual df | 6,319 | ||
| No. Obs. | 6,324 | ||
| 1 CI = Confidence Interval | |||
set_gtsummary_theme
gtsummaryhas a number of themes that allow us to format the tables for different journalsSome available formats are:
theme_jama()theme_lancet()theme_bmj()theme_annals()
Here we format to the JAMA journal format
Compare model results
We might want to combinete several different model results into a single table.
First we format each of the models using
tbl_regression()
# Note for this first one that I am showing how to integrate this
# into a workflow where you start with the analytic data frame,
# pipe it into lm() and then pipe the results into
# tbl_regression.
# BUT: note that the first pipe has to be the magrittr pipe %>%
# and not the "new" pipe |>
tbl_lm_model1 <- df_completecase %>%
lm(BPSysAve ~ factor(Education),
data=.) |>
tbl_regression(intercept=TRUE,
label = list('factor(Education)' ~ 'Education'))Compare model results
tbl_lm_model2 <- lm_model2 |>
tbl_regression(intercept=TRUE,
label = list('factor(Education)' ~ 'Education',
'factor(agecat)' ~ 'Age category'))
tbl_lm_model3 <- lm_model3 |>
tbl_regression(intercept=TRUE,
label = list('factor(Education)' ~ 'Education',
'factor(agecat)' ~ 'Age category',
'factor(racecat)' ~ 'Racialized group'))
tbl_lm_model4 <- lm_model4b |>
tbl_regression(intercept=TRUE,
label = list('factor(Education)' ~ 'Education',
'factor(agecat)' ~ 'Age category',
'interaction(Gender, factor(racecat))' ~ 'Gender X Racialized group'),
)Compare model results
- Now that each of the models has been formatted, I can use
tbl_mergeto put the models together to be shown side-by-side
Compare model results
| Characteristic | Model 1 | Model 2 | Model 3 | Model 4 | ||||
|---|---|---|---|---|---|---|---|---|
| Beta (95% CI)1 | p-value | Beta (95% CI)1 | p-value | Beta (95% CI)1 | p-value | Beta (95% CI)1 | p-value | |
| (Intercept) | 119 (118 to 119) | <0.001 | 109 (108 to 110) | <0.001 | 109 (108 to 110) | <0.001 | 109 (108 to 110) | <0.001 |
| Education | ||||||||
| College Grad | — | — | — | — | ||||
| Some College | 3.8 (2.7 to 4.8) | <0.001 | 3.2 (2.2 to 4.2) | <0.001 | 2.9 (1.9 to 3.9) | <0.001 | 2.9 (1.9 to 3.9) | <0.001 |
| High School | 5.1 (3.9 to 6.4) | <0.001 | 3.9 (2.7 to 5.0) | <0.001 | 3.4 (2.3 to 4.5) | <0.001 | 3.4 (2.3 to 4.5) | <0.001 |
| 9 - 11th Grade | 4.7 (3.3 to 6.1) | <0.001 | 3.2 (1.9 to 4.5) | <0.001 | 2.7 (1.3 to 4.0) | <0.001 | 2.7 (1.3 to 4.0) | <0.001 |
| 8th Grade | 7.9 (6.1 to 9.8) | <0.001 | 3.7 (1.9 to 5.4) | <0.001 | 3.6 (1.8 to 5.5) | <0.001 | 3.7 (1.9 to 5.5) | <0.001 |
| Age category | ||||||||
| 25-34 | — | — | — | |||||
| 35-44 | 4.0 (2.8 to 5.2) | <0.001 | 4.1 (2.9 to 5.3) | <0.001 | 4.1 (2.8 to 5.3) | <0.001 | ||
| 45-54 | 6.7 (5.5 to 7.9) | <0.001 | 6.7 (5.5 to 7.9) | <0.001 | 6.6 (5.4 to 7.8) | <0.001 | ||
| 55-64 | 13 (11 to 14) | <0.001 | 13 (11 to 14) | <0.001 | 13 (11 to 14) | <0.001 | ||
| 65-74 | 16 (14 to 17) | <0.001 | 16 (14 to 17) | <0.001 | 16 (14 to 17) | <0.001 | ||
| 75+ | 24 (22 to 25) | <0.001 | 24 (22 to 25) | <0.001 | 24 (22 to 25) | <0.001 | ||
| Gender | ||||||||
| female | — | — | ||||||
| male | 4.0 (3.2 to 4.8) | <0.001 | 4.1 (3.3 to 4.8) | <0.001 | ||||
| Racialized group | ||||||||
| White Non-Hispanic | — | |||||||
| Black Non-Hispanic | 4.4 (3.1 to 5.7) | <0.001 | ||||||
| Hispanic | 0.03 (-1.2 to 1.3) | 0.96 | ||||||
| Other Non-Hispanic | -1.3 (-2.8 to 0.24) | 0.10 | ||||||
| Gender X Racialized group | ||||||||
| female.White Non-Hispanic | — | |||||||
| male.White Non-Hispanic | 3.7 (2.8 to 4.6) | <0.001 | ||||||
| female.Black Non-Hispanic | 4.8 (3.0 to 6.6) | <0.001 | ||||||
| male.Black Non-Hispanic | 7.7 (5.9 to 9.6) | <0.001 | ||||||
| female.Hispanic | -0.72 (-2.5 to 1.1) | 0.43 | ||||||
| male.Hispanic | 4.4 (2.7 to 6.1) | <0.001 | ||||||
| female.Other Non-Hispanic | -3.0 (-5.1 to -0.92) | 0.005 | ||||||
| male.Other Non-Hispanic | 4.3 (2.1 to 6.5) | <0.001 | ||||||
| 1 CI = Confidence Interval | ||||||||
Output options
- In addition to embedding in a RMarkdown or Quarto file, we can also output directly to an html file
- We can save to a Word file (docx)
- We can save to an Excel file
A workflow for plotting model estimates
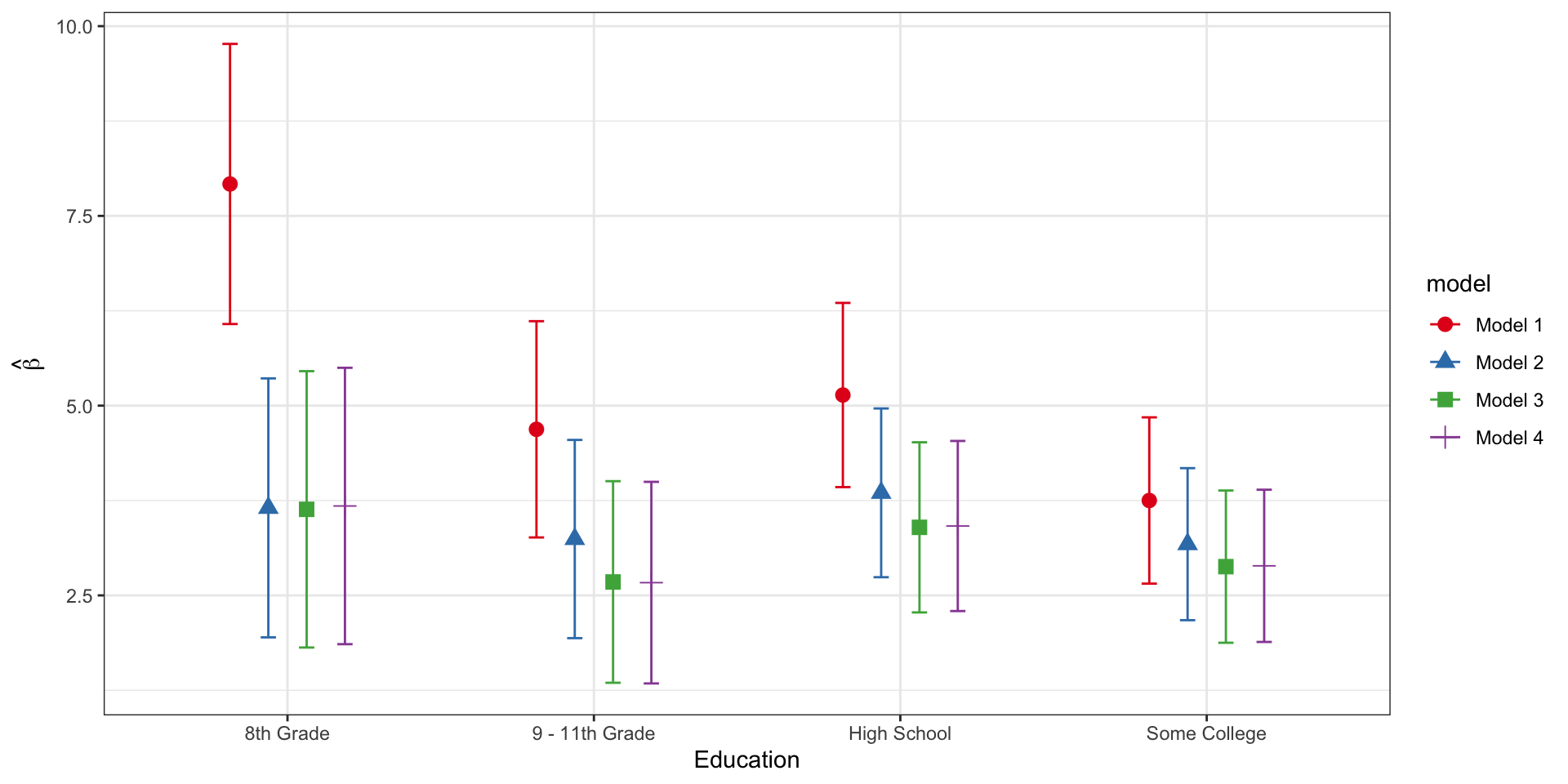
Let’s say that we want to compare estimates of the education effect in the crude and adjusted models.
Here, I show an example of using
broom::tidyto- extract the model estimates,
- stack them together in a tibble,
- filter out just the education terms,
- and pipe the tibble into
ggplotin order to plot the estimates.
.{smaller}
# Extract the education effects from each model and combine in a tibble
lm_education_estimates <- bind_rows(broom::tidy(lm_model1, conf.int=TRUE) %>%
mutate(model = "Model 1"),
broom::tidy(lm_model2, conf.int=TRUE) %>%
mutate(model = "Model 2"),
broom::tidy(lm_model3, conf.int=TRUE) %>%
mutate(model = "Model 3"),
broom::tidy(lm_model4b, conf.int=TRUE) %>%
mutate(model = "Model 4")) %>%
# here, we use stringr::str_detect to detect the entries
# where term includes the string 'Education'
filter(stringr::str_detect(term, "Education")) %>%
# here, we use the separate() function to pull out the category labels
# from term so that we can have nice labeling in the plot
separate(col=term, sep=17, into=c("term", "category"), convert=TRUE)# Use ggplot to plot the point estimates and 95% CIs
# Note that we are differentiating the models by color AND by the shape of the plotting symbol
ggplot(lm_education_estimates, aes(x=category, y=estimate, color=model, shape=model)) +
# position=position_dodge() is specified so that the estimates are side by side rather than
# plotted on top of one another
geom_point(position=position_dodge(0.5), size=3) +
# geom_errorbar allows us to plot the 95% confidence limits
geom_errorbar(aes(ymin=conf.low, ymax=conf.high), position=position_dodge(0.5), width=0.2) +
# scale_color_brewer allows me to control the colors for plotting the different models
scale_color_brewer(palette="Set1") +
labs(x="Education", y=expression(hat(beta))) +
theme_bw()